For IRD iTrop or IFB HPC resources users ONLY
# On i-Trop HPC
module load system/singularity/3.3.0
module load system/python/3.7.2
# On IFB HPC
module load singularity
module load python/3.7
module load graphviz/2.40.
Prepare config.yaml¶
To run the pipeline you have to provide the data path and activate/deactivate options in every step from config.yaml file.
1. Providing data¶
First, indicate the data path in the configuration file
DATA:
FASTQ: '/path/to/fastq/directory/'
REF: '/path/to/referencefile.fasta'
GENOME_SIZE: '1m'
FAST5: '/path/to/fast5/directory/'
ILLUMINA: '/path/to/illumina/directory/'
OUTPUT: '/path/to/output/directory/'
CIRCULAR : True/False
- FASTQ: CulebrONT takes as input a FASTQ directory. Every FASTQ file should contain the whole set of reads to be assembled (meaning that multiple runs must be merged in a single FASTQ file), as each FASTQ file found in this repertory will be assembled independently. FASTQ files can be compressed or not (gzipped). Naming convention accepted by CulebrONT are: NAME.fastq.gz or NAME.fq.gz or NAME.fastq or NAME.fq.
- REF: Only one REFERENCE genome file will be used by CulebrONT. This REFERENCE will be used for QC steps (QUAST and MAUVE).
- GENOME_SIZE : Estimated genome size (m,g,k) of the assembly.
- FAST5: Nanopolish needs FAST5 files to training steps. Please give the path of FAST5 repertory in the FAST5 DATA parameter. Inside this directory, a subdirectory with the exact same name as the corresponding FASTQ (before the .fastq.gz) is requested. For instance, if in the FASTQ directory we have run1.fastq.gz and run2.fastq.gz, CulebrONT is expecting the run1/ and run2/ subdirectories in the FAST5 main directory.
- ILLUMINA : indicate the path to the directory with Illumina sequence data (in fastq or fastq.gz format) to perform KAT quality. Use preferentially paired-end data.
- OUTPUT: output path directory.
- CIRCULAR : Indicate True or False to activate/deactivate circularisation steps (only to procaryote).
2. Chose assemblers, polisher and correctors¶
Activate/deactivate assemblers and correctors as you wish. By default, Racon is launched as POLISH tool after each activated assembly step. You must activate at least one assembler and one corrector.
Example:
ASSEMBLY:
CANU : False
FLYE : True
MINIASM : False
SHASTA : False
SMARDENOVO : True
RAVEN: True
CORRECTION:
NANOPOLISH : True
MEDAKA : False
3. Chose quality tools¶
Activate/deactivate quality tools as you wish. By default, BUSCO AND QUAST are launched if ‘True’ in QUALITY (ASSEMBLY OR POLISHING OR CORRECTION OR both) steps.
You must to activate at least one QUALITY step.
Example:
QUALITY:
ASSEMBLY : True
POLISHING : True
CORRECTION : True
Others quality tools are launched only on the final assemblies (BLOBTOOLS, ASSEMBLYTICS, WEESAM and KAT).
KAT quality tool can be activate but Illumina reads are mandatory in this case.
#### Others quality tools
WEESAM: True
BLOBTOOLS: True
ASSEMBLYTICS: True
KAT: True
Alignment of various assemblies for small genomes (<10-20Mbp) is also possible by using Mauve. Mauve will compared each state of the assembly (Raw, Polished and Corrected) for each assembler used.
A Fixstart step is possible before Mauve MSA to improve alignment on circular molecules.
- Fixstart will be deactivated if CIRCULAR is False
- Only activate MAUVE if you have more than one sample and more than one quality step.
MSA:
FIXSTART: True
MAUVE: True
4. Parameters for some specific tools¶
Specifically to Racon:
- Racon can be launch recursively from 1 to 9 rounds. 2 or 3 are recommanded.
Specifically to Medaka :
- If ‘MEDAKA_TRAIN_WITH_REF’ is activated, Medaka launchs training using the reference found in ‘DATA/REF’ param. Medaka does not take into account other medaka model parameters.
- If ‘MEDAKA_TRAIN_WITH_REF’ is deactivated, Medaka does not launch training but uses instead the model provided in ‘MEDAKA_MODEL_PATH’. If ‘MEDAKA_MODEL_PATH’ is empty, this param is not used and the default model for E.coli is used.
Standard parameters used:
############ PARAMS ################
params:
MINIMAP2:
PRESET_OPTION: 'map-pb' # -x minimap2 preset option is map-pb by default (map-pb, map-ont etc)
CANU:
MAX_MEMORY: '15G'
OPTIONS: '-fast'
SMARTDENOVO:
KMER_SIZE: '16'
OPTIONS: '-J 5000'
SHASTA:
MEM_MODE: 'filesystem'
MEM_BACKING: 'disk'
CIRCLATOR:
OPTIONS: ''
RACON:
RACON_ROUNDS: 2 #1 to 9
NANOPOLISH:
# segment length to split assembly and correct it default=50000
NANOPOLISH_SEGMENT_LEN: '50000'
# overlap length between segments default=200
NANOPOLISH_OVERLAP_LEN: '200'
OPTIONS: ''
MEDAKA:
# if 'MEDAKA_TRAIN_WITH_REF' is True, Medaka launchs training using the reference found in DATA REF param. Medaka does not take in count other Medaka model parameters below.
MEDAKA_TRAIN_WITH_REF: True
MEDAKA_MODEL_PATH: 'medakamodel/r941_min_high_g303_model.hdf5' # if empty this param is not used.
BUSCO:
DATABASE : 'Data-Xoo-sub/bacteria_odb10'
MODEL : 'genome'
# 'SP' : 'caenorhabditis'
SP : ''
QUAST:
REF: 'Data-Xoo-sub/ref/BAI3_Sanger.fsa'
GFF: ''
GENOME_SIZE_PB: 48000000
#GENOME_SIZE_PB: 1000000
OPTIONS : ''
DIAMOND:
DATABASE: 'Data-Xoo-sub/testBacteria.dmnd'
MUMMER:
# -l default 20
MINMATCH : 100
# -c default 65
MINCLUSTER: 500
ASSEMBLYTICS:
UNIQUE_ANCHOR_LEN: 10000
MIN_VARIANT_SIZE: 50
MAX_VARIANT_SIZE: 10000
Singularity containers¶
To use Singularity containers, provide to CulebrONT the already build Singularity containers path on your computer or cluster.
As an example, here are singularity images found on the i-Trop HPC from the SouthGreen platform.
# cluster with scratch temporary directory
SCRATCH = False
## @ITROP PATH
tools:
## ASSEMBLERS:
CANU_SIMG : '/data3/projects/containers/CULEBRONT/canu-1.9.simg'
FLYE_SIMG : '/data3/projects/containers/CULEBRONT/flye-2.7.1.simg'
MINIASM_SIMG : '/data3/projects/containers/CULEBRONT/miniasm-0.3.simg'
MINIPOLISH_SIMG : '/data3/projects/containers/CULEBRONT/minipolish-0.1.2.simg'
RAVEN_SIMG : '/data3/projects/containers/CULEBRONT/raven_conda-gpu-v1.1.10.simg'
SMARTDENOVO_SIMG : '/data3/projects/containers/CULEBRONT/smartdenovo.simg'
SHASTA_SIMG : '/data3/projects/containers/CULEBRONT/shasta-0.5.1.simg'
## CIRCULARISATION
CIRCLATOR_SIMG : '/data3/projects/containers/CULEBRONT/circlator-1.5.5.simg'
## POLISHERS:
RACON_SIMG : '/data3/projects/containers/CULEBRONT/racon-1.4.3.simg'
NANOPOLISH_SIMG : '/data3/projects/containers/CULEBRONT/nanopolish-0.11.3.simg'
## CORRECTION
MEDAKA_SIMG : '/data3/projects/containers/CULEBRONT/medaka_conda-gpu-1.0.3.simg'
## QUALITY
BUSCO_SIMG : '/data3/projects/containers/CULEBRONT/busco-4.0.5.simg'
QUAST_SIMG : '/data3/projects/containers/CULEBRONT/quast-5.0.2.simg'
WEESAM_SIMG : '/data3/projects/containers/CULEBRONT/weesam.simg'
BLOBTOOLS_SIMG : '/data3/projects/containers/CULEBRONT/bloobtools-v1.1.1.simg'
MINIMAP2_SIMG: '/data3/projects/containers/CULEBRONT/nanopolish-0.11.3.simg'
DIAMOND_SIMG : '/data3/projects/containers/CULEBRONT/diamond-0.9.30.simg'
MUMMER_SIMG : '/data3/projects/containers/CULEBRONT/mummer-4beta.simg'
ASSEMBLYTICS_SIMG : '/data3/projects/containers/CULEBRONT/assemblytics-1.2.simg'
SAMTOOLS_SIMG : '/data3/projects/containers/CULEBRONT/nanopolish-0.11.3.simg'
KAT_SIMG : '/data3/projects/containers/CULEBRONT/kat-2.4.2.simg'
MINICONDA_SIMG : 'shub://vibaotram/singularity-container:cpu-guppy3.4-conda-api'
R_SIMG: '/data3/projects/containers/CULEBRONT/R.simg'
Available recipes from containers are available in the Containers folder, as well as on the main CulebrONT repository. Feel free to build them on your own computer (or cluster); be careful, you need root rights to do it.
Built singularity images are also available on https://itrop.ird.fr/culebront_utilities/. Feel free to download it using wget for example.
singularity hub¶
If you want to recover singularity images from the Singularity Hub and build them, please use these paths :
# cluster with scratch temporal repertory
SCRATCH : False
tools:
###### ASSEMBLERS:
CANU_SIMG: 'shub://SouthGreenPlatform/CulebrONT_pipeline:canu-1.9.def'
FLYE_SIMG: 'shub://SouthGreenPlatform/CulebrONT_pipeline:flye-2.6.def'
MINIASM_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:miniasm-0.3.def'
MINIPOLISH_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:minipolish-0.1.2.def'
RAVEN_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:raven_conda-gpu-v1.1.10.simg'
SMARTDENOVO_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:smartdenovo.simg'
SHASTA_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:shasta-0.5.1.simg'
###### CIRCULARISATION
CIRCLATOR_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:circlator-1.5.5.def'
###### POLISHERS:
RACON_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:racon-1.4.3.def'
###### CORRECTION
NANOPOLISH_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:nanopolish-0.11.3.def'
MEDAKA_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:medaka_conda-gpu-1.0.3.simg'
###### QUALITY
BUSCO_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:busco-4.def'
QUAST_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
WEESAM_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
BLOBTOOLS_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
MINIMAP2_SIMG: 'shub://SouthGreenPlatform/CulebrONT_pipeline:nanopolish-0.11.3.simg'
DIAMOND_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
MUMMER_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:mummer-4beta.def'
ASSEMBLYTICS_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
SAMTOOLS_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:nanopolish-0.11.3.simg'
KAT_SIMG : 'shub://SouthGreenPlatform/CulebrONT_pipeline:quality.def'
MINICONDA_SIMG: 'shub://vibaotram/singularity-container:cpu-guppy3.4-conda-api'
R_SIMG: 'shub://SouthGreenPlatform/CulebrONT_pipeline:r.def'
Launching on a single machine¶
To launch CulebrONT on a single machine, you should use the parameters --use-singularity and --use-conda.
See the example below:
Launching on HPC clusters¶
Preparing Slurm cluster configuration using cluster_config.yaml¶
On cluster_config.yaml, you can add partition, memory and threads to be used by default for each rule. If more memory or threads are requested, please adapt the content of this file before running on a cluster for every rule. For instance give more memory to Canu and Medaka.
Here is a example of the configuration file we used on the i-Trop HPC.
__default__:
cpus-per-task : 4
ntasks : 1
mem-per-cpu : '2'
partition : "normal"
output : 'logs/stdout/{rule}/{wildcards}'
error : 'logs/error/{rule}/{wildcards}'
#
#run_nanopolish :
# cpus-per-task : 8
# mem-per-cpu : '4'
# partition : "long"
#
run_canu:
cpus-per-task : 8
mem-per-cpu : '4'
partition : "long"
submit_culebront.sh¶
This is a typical launcher for using CulebrONT on a Slurm cluster. You have to adapt it for the configuration of your favorite cluster. Please adapt this script also if you want to use wrappers or profiles.
This launcher can be submitted to the Slurm queue typing:
Important : Do not forget to adapt submit_culebront.sh if you want to use wrappers or profile!!
slurm_wrapper¶
A slurm_wrapper.py script is available on CulebrONT projet to manage ressources from your cluster configuration (taken from cluster_config.yaml file). This is the easier way to know what is running on cluster and to adapt ressources for every job. Take care, this cluster_config.yaml file is becomming obsolete on next Snakemake versions.
Profiles¶
Optionally is possible to use Profiles in order to run CulebrONT on HPC cluster. Please follow the recommandations found on the SnakeMake profile github.
Here is an example of how to profile a Slurm scheduler using those recommandations.
SLURM Profile slurm-culebrONT is now created on : /shared/home/$USER/.config/snakemake/slurm-culebrONT repertory
Output on CulebrONT¶
The architecture of CulebrONT output is designed as follows:
The same Architecture per sample (fastq = SAMPLE-1 in example) is followed for LOG files.
Report¶
CulebrONT generates a beautiful report containing, foreach fastq found on input directory, a summary of interesting statistics such as busco, quast and others ones that you will discover!
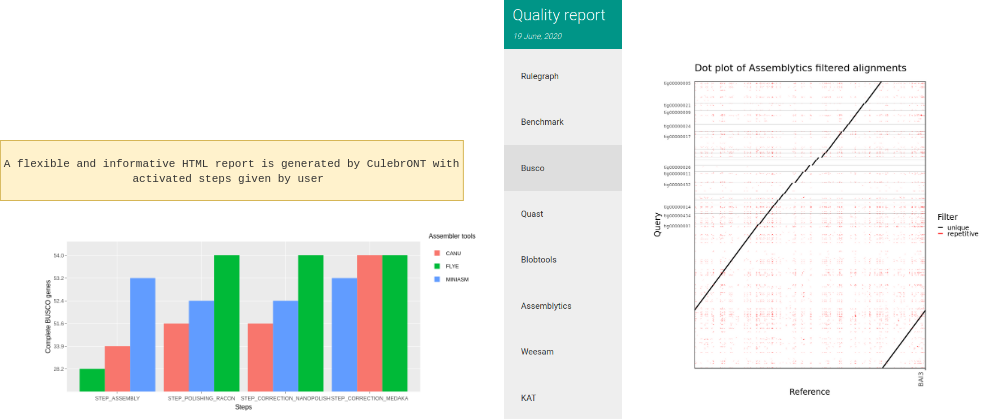
Important: To visualise the report created by CulebrONT, transfer the whole REPORT directory on your local machine before opening the report.html file with a navigator.